Parameter Estimation and Ensemble Modeling
Current Participants: Yang Liu , Sandro Hutter
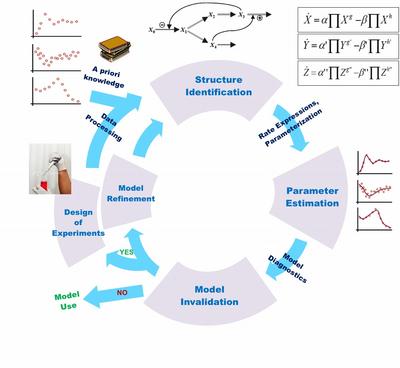
Biological system model identification is typically formulated as an iterative process that integrates wet-lab experiments and in silico analysis and optimization (see Figure 1). Most biological modeling studies in the literature involve the creation of an accurate (high fidelity) model of the system, defined by a set of equations and parameter values that describe important or interesting behavior of the system. There are many challenges in applying this iterative model identification procedure to a real-life problem. The bottlenecking step is typically encountered during the estimation of unknown kinetic parameters from experimental data. This has led to the development of a large number of parameter estimation techniques. However, as we and others have shown, the estimation of kinetic parameters by fitting model simulations to biological data is usually ill-posed. There often does not exist a single (best-fit) solution to the data fitting problem, but rather one can find many parameter combinations, i.e. an ensemble of parameters that can fit the data statistically equally well. Consequently, the best-fit model, even if one is obtained, may have little predictive capability, or worse, it could be misleading.
In light of the identifiability issue and other uncertainty, the success of any model-based studies that rely on building an accurate model of the system, will depend on balancing the (numerical and experimental) tractability of model identification and model fidelity. In this project, we take a different approach, based on the realization that in a real-life problem, the model itself is not an end, but rather a means toward a goal (e.g., obtaining biological insights through model analysis or optimizing the production of certain biomolecules). Specifically, we are using an ensemble modeling approach. The ensemble consists of models that cannot be differentiated among each other from available information about the system (prior knowledge, experimental data). Consequently, the ensemble directly represents the uncertainty about the system due to incomplete information. Our research in this project has generated several tools for parameter estimation and ensemble modeling, which are available through an easy-to-use MATLAB interface called REDEMPTION.
References
- Liu, Y. and Gunawan, R. Bioprocess optimization under uncertainty using ensemble modeling. 2017. J. Biotechnology. Accepted.
- Liu, Y., Manesso, E. and Gunawan, R. REDEMPTION: Reduced Dimension Ensemble Modeling and Parameter Estimation. Bioinformatics (2015). external page abstract
- Liu, Y., and Gunawan, R. Parameter estimation of dynamic biological network models using integrated fluxes. BMC Systems Biology 8, 127 (2014). external page abstract
- Jia, G., Stephanopoulos, G., Gunawan, R. Incremental parameter estimation of kinetic metabolic network models. BMC Systems Biology 6, 142 (2012). external page abstract
- Jia, G., Stephanopoulos, G. & Gunawan, R. Ensemble Kinetic Modeling of Metabolic Networks from Dynamic Metabolic Profiles. Metabolites 2, 891–912 (2012). external page abstract
- Jia, G., Stephanopoulos, G. N. & Gunawan, R. Parameter estimation of kinetic models from metabolic profiles : two-phase dynamic decoupling method. Bioinformatics 27, 1964–1970 (2011). external page abstract